These algorithms are used and described in:
Lee et al.: “Transfer Entropy Estimation and Directional Coupling Change Detection in Biomedical Time Series.” Biomedical Engineering Online 2012 11:19.Please cite the above publication when referencing this material, and also include the standard citation for PhysioNet: Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. "PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals," Circulation 101(23):e215-e220 [Circulation Electronic Pages; http://circ.ahajournals.org/content/101/23/e215.full]; 2000 (June 13).
Description
This is a repository of MATLAB functions that can estimate transfer entropy (information flow) from one time series to another using a non-parametric partitioning algorithm. Also included is an example data set that the implemented algorithms can be applied to.
The functions have been tested in MATLAB R2016b on 03 March 2016.
Usage
There are three functions used to calculate transfer entropy between time series using different methods of probability density estimation:
- [T] = transferEntropyKDE(X,Y,t,w,N,bw_coeff)
- Based on Guassian kernel density estimation.
- Calls function mdKDE
- [T nPar dimPar]=transferEntropyPartition(X,Y,t,w)
- Based on the Darbellay-Vajda partitioning algorithm
- Calls function DVpartition3D
- [T] = transferEntropyRank(X,Y,l,k,t,w,Q)
- Based on bin counting with fixed and equally-spaced bins.
- Calls function quantentr
The MAT file 'example_data.mat' contains a 1-D structure array of example data that harnesses an information flow from X to Y at a lag of 2 according to: y(i)=[(1+a)*x(i-2)]^2
Different values of the coupling constant 'a' were simulated, along with associated levels of Laplacian noise given by SNR levels.
- For a demonstration, run 'demoscript.m'. The plotted results should look like figure 1.
- See the individual .m function files for information on how to call them.
- See readme_example_data.txt for information on the example data parameters.
- See Measuring Information Transfer, Physical Review Letters, 85(2):461-464, 2000 for more details on the methods.
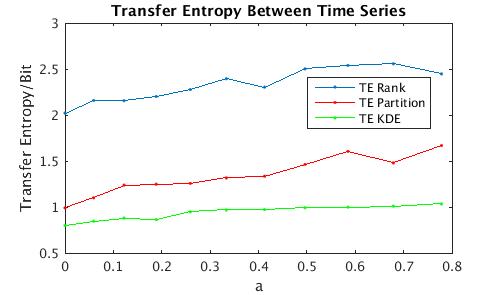
Figure 1: Transfer entropy between time series with different coupling constants.Comparison of all 3 algorithms.
Acknowledgments
This package was developed by J. Lee, S. Nemati, and I. Silvia.
Name Last modified Size Description
Parent Directory -
transferEntropyRank.m 04-Mar-2016 01:22 2.4K
transferEntropyPartition.m 04-Mar-2016 01:22 2.4K
transferEntropyKDE.m 04-Mar-2016 01:22 2.5K
readme_example_data.txt 10-Jun-2016 18:50 703
quantentr.m 04-Mar-2016 01:22 1.3K
mdKDE.m 04-Mar-2016 01:22 1.6K
local.css 04-Mar-2016 00:13 3.1K C# source file
example_data.mat 04-Mar-2016 01:22 84K
demoscript.m 04-Mar-2016 01:22 606
demoresults.jpg 04-Mar-2016 01:56 18K
DVpartition3D.m 04-Mar-2016 01:22 3.7K
COPYING 07-Apr-2016 01:05 34K GNU General Public License
|
If you would like help understanding, using, or downloading content, please see our Frequently Asked Questions. If you have any comments, feedback, or particular questions regarding this page, please send them to the webmaster. Comments and issues can also be raised on PhysioNet's GitHub page. Updated Friday, 28-Oct-2016 22:58:42 CEST |
PhysioNet is supported by the National Institute of General Medical Sciences (NIGMS) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) under NIH grant number 2R01GM104987-09.
|