This database is described in
Please cite this publication when referencing this material, and also include the standard citation for PhysioNet:
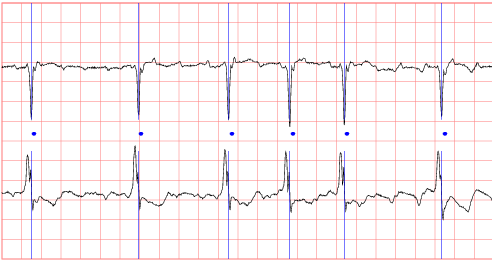
This database of two-channel ECG recordings has been created for use in the Computers in Cardiology Challenge 2004, an open competition with the goal of developing automated methods for predicting spontaneous termination of atrial fibrillation (AF). See the challenge announcement for information about the competition.
The database is divided into a learning set (records with names of the form n*, s*, and t*) and two test sets (records with names of the form a* and b*).
If you wish to download all of the files in this directory without selecting each one individually, try using a utility for batch HTTP transfers such as wget, available here in source form for all versions of UNIX and as a precompiled binary for MS-Windows. Most Linux distributions include wget. Once you have installed wget, retrieve these files using
wget -r -np http://www.physionet.org/physiobank/database/aftdb/
(or substitute the name of a nearby PhysioNet mirror for www.physionet.org above). The files in this database occupy about 3.3 megabytes in all.
Each record is a one-minute segment of atrial fibrillation, containing two ECG signals, each sampled at 128 samples per second. The segments were extracted from long-term (20-24 hour) ECG recordings. Each record includes a set of QRS annotations produced by an automated detector, in which all detected beats, including any ectopic beats, are labelled as normal. These annotations have not been audited and may contain a small number of errors.
The learning set contains 30 records in all, with 10 records in each of three groups:
- Group N (records n01, n02, ... n10): non-terminating AF (defined as AF that was not observed to have terminated for the duration of the long-term recording, at least an hour following the segment).
- Group S (records s01, s02, ... s10) AF that terminates one minute after the end of the record.
- Group T (records t01, t02, ... t10) AF that terminates immediately (within one second) after the end of the record. Note that these records come from the same long-term ECG recordings as those in group S and immediately follow the Group S records (for example, t01 is the continuation of s01).
The learning set records were obtained from 20 different subjects (10 group N, 10 group S/T).
Test set A contains 30 records (a01, a02, ... a30) from 30 subjects (none represented in the learning set or in test set B). Approximately half of these records belong to group N, and the others belong to group T. The goal of the first challenge event is to identify which records in test set A belong to group T.
Test set B contains 20 records (b01, b02, ... b20), 2 from each of 10 subjects (none represented in the learning set or in test set A). One record of each pair belongs to group S, and the other to group T; note, however, that there are short gaps (of less than one second) between some of these pairs. The goal of the second challenge event is to identify which records belong to group T.
Several files are associated with each record. The files with names of the form *.dat contain the digitized ECGs (16 bits per sample, least significant byte first in each pair, 128 samples per signal per second, samples from each channel alternating, nominally 200 A/D units per millivolt). The .hea files are (text) header files that specify the names and formats of the associated signal files; these header files are needed by the software available from this site. The .qrs files are machine-generated (binary) annotation files, provided for the convenience of those who do not wish to use their own QRS detectors. Please note that the .qrs files are unaudited and contain errors. You may wish to correct these errors (if you do, please send your corrections to us). Otherwise, you may use these annotations in uncorrected form if you wish to investigate methods of predicting AF terminationthat are robust with respect to small numbers of QRS detection errors, or you may ignore these annotations entirely and work directly from the signal files.
You may wish to begin your exploration of this database using the Chart-O-Matic, which allows you to view any of these records using your web browser (no special software is required). PhysioToolkit includes a large amount of free software that may be useful for studying these records further.
Acknowledgements
Special thanks to Steven Swiryn of Northwestern University, who provided the recordings excerpted here.
Name Last modified Size Description
Parent Directory -
ANNOTATORS 23-Jun-2010 23:59 31 list of annotators
DOI 21-Sep-2015 19:00 19
MD5SUMS 13-Jul-2005 02:30 13K
RECORDS 23-Jun-2010 23:59 1.2K list of record names
SHA1SUMS 13-Jul-2005 02:30 15K
SHA256SUMS 18-Sep-2007 18:36 378
learning-set/ 17-Sep-2003 09:46 -
local.css 10-Nov-2015 21:16 3.1K
test-set-a/ 09-Mar-2016 20:43 -
test-set-b/ 17-Sep-2003 09:47 -
|
If you would like help understanding, using, or downloading content, please see our Frequently Asked Questions. If you have any comments, feedback, or particular questions regarding this page, please send them to the webmaster. Comments and issues can also be raised on PhysioNet's GitHub page. Updated Friday, 28-Oct-2016 22:58:42 CEST |
PhysioNet is supported by the National Institute of General Medical Sciences (NIGMS) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) under NIH grant number 2R01GM104987-09.
|