


Next: A Research Example
Up: Software Execution
Previous: Viewing Cardiac Function and Venous Return Curves
Viewing Examples
The following examples illustrate how to view waveforms and cardiac
function/venous return curves on-line and off-line. Prior to
implementing these examples, the researcher should set the time
duration displayed by the WAVE window to 20 seconds by resizing the
window and/or adjusting the Time scale variable (click VIEW option at
the top of the WAVE menu bar).
Ex. 1
- Desired Execution:
- On-line display of left ventricle pressure, volume, and flow
rate.
- Uncontrolled, unperturbed, intact pulsatile heart and
circulation with default parameter values.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_1.
- Open the file parameters_1 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: waveform: 0 9 17 and
annotations: 0. Make sure all of the status parameters are
set to zero.
- Save the file parameters_1.
- Run the following command at the Linux prompt:
rcvsim parameters_1 foo1
- Execution Output:
- The WAVE window in Figure 10 will initially appear
and will automatically scroll through the simulated data as they are
being generated. This process will terminate once 300 seconds of
the data have been simulated.
- The following files will be created in the current directory:
foo1.dat, foo1.qrs, foo1.hea, and parameters_1.0 which may subsequently be viewed off-line (See
Example 2).
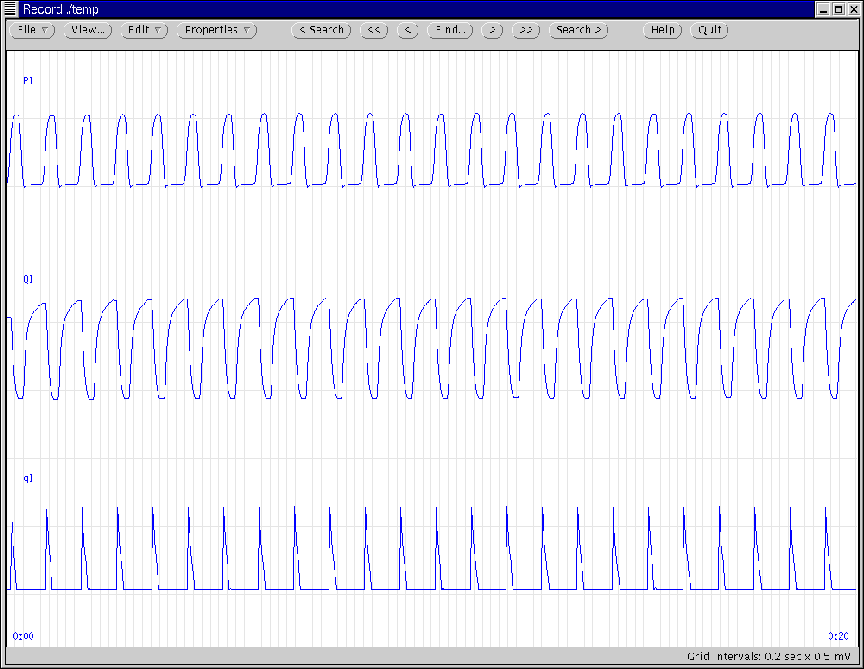
Figure 10:
Initial WAVE window generated according to Ex. 1
and Ex. 2.
 |
Ex. 2
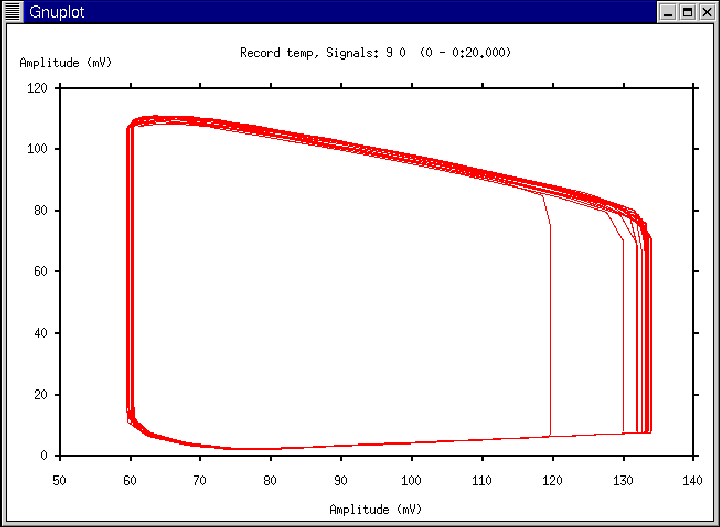
Figure 11:
Gnuplot window generated according to Ex. 2.
 |
Ex. 3
- Desired Execution:
- On-line display of systemic arterial pressure, heart rate, and
instantaneous lung volume with annotations.
- Fully controlled, fully perturbed (fixed-rate breathing), intact
pulsatile heart and circulation with default parameter values.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_3.
- Open the file parameters_3 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: waveform: 1 15 26,
baro: 3, dncm: 1, breathing: 1, dra: 1, and
df: 1.
- Save the file parameters_3.
- Execute the following command at the Linux prompt:
rcvsim parameters_3 foo3
- Execution Output:
- A WAVE window will appear and will automatically scroll through
the simulated data with annotations. Figure 12
illustrates the window after one minute of data has been calculated.
This process will terminate once 300 seconds of data have been
simulated.
- The following files will be created in the current directory:
foo3.dat, foo3.qrs, foo3.hea, and parameters_3.0.
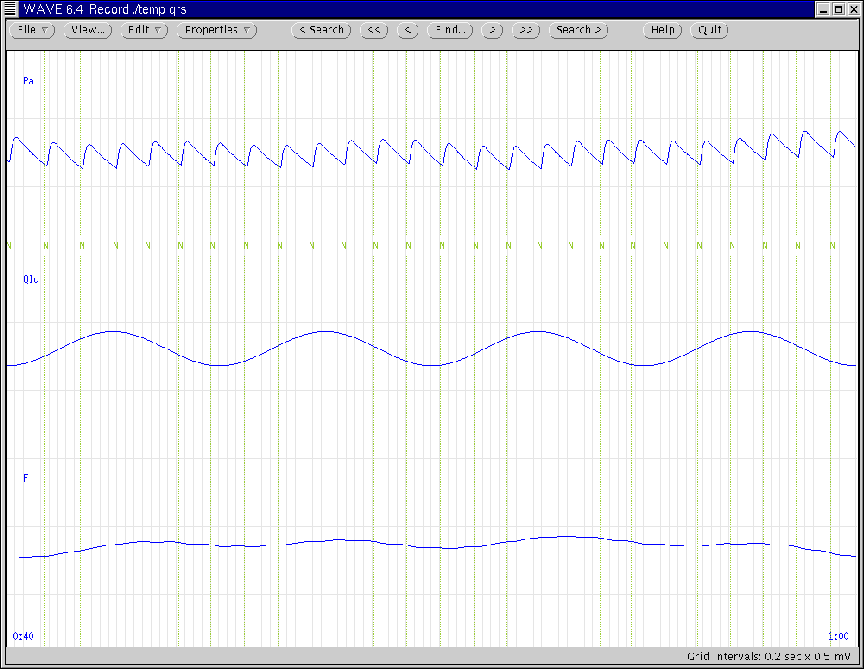
Figure 12:
WAVE window generated according to Ex. 3 and Ex. 4.
 |
Ex. 4
Ex. 5
- Desired Execution:
- On-line display of systemic arterial pressure and volume with
annotations.
- Uncontrolled, unperturbed, intact pulsatile heart and
circulation initially with default parameter values.
- On-line reduction in systemic arterial compliance by a factor of
two.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_5.
- Open the file parameters_5 with any text editor (e.g.,
emacs).
- Re-assign the following parameter: waveform: 1 10. Make
sure all of the status parameters are assigned the numerical value
of zero.
- Save the file parameters_5.
- Run the following command at the Linux prompt:
rcvsim parameters_5 foo5
- Some time in the midst of the simulation, type ``p'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Re-assign the the following parameter: Ca: 0.8.
- Save the file parameters_5.
- Type ``r'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Execution Output:
- A WAVE window will appear and will automatically scroll through
the simulated data with annotations. The automatic scrolling will
stop once Step 6. is executed. (At this point, the researcher may
scroll backwards with the the arrow buttons at the top of the WAVE
display system.) After Steps 7.-9. are executed, automatic
scrolling will resume until a total of 300 seconds of data have been
calculated. Figure 13 illustrates the WAVE window
during the time of reduction in systemic arterial compliance. Note
that this reduction is annotated with the name of the saved
parameter file. (Also note how systemic arterial volume is
conserved through the instantaneous change in systemic arterial
pressure.)
- The following files will be created in the current directory:
foo5.dat, foo5.qrs, foo5.hea, parameters_5.0,
and parameters_5.1.
Figure 13:
WAVE window generated according to Ex. 5.
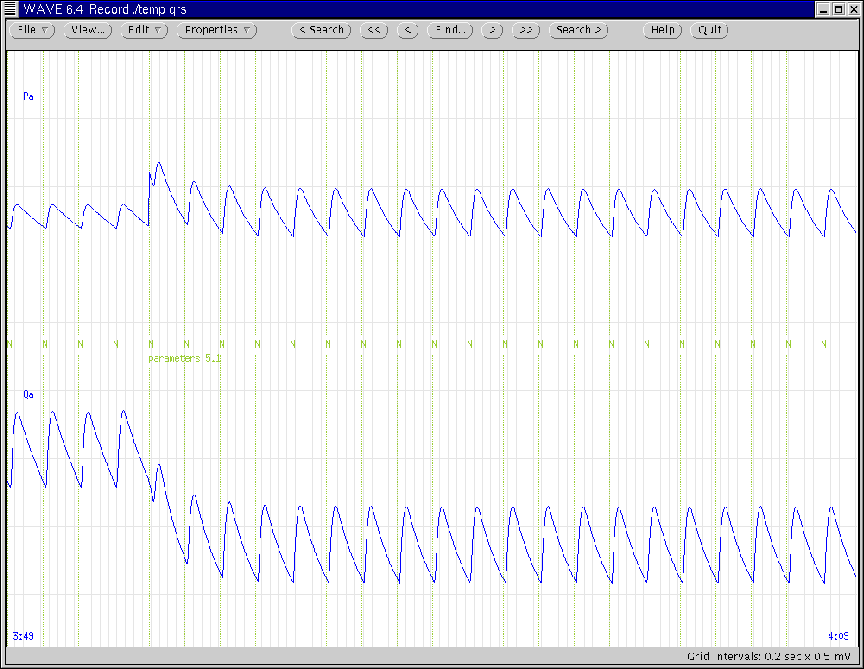 |
Ex. 6
- Desired Execution:
- On-line display of systemic arterial pressure, intrathoracic
pressure, and instantaneous lung volume with annotations.
- Uncontrolled, intact pulsatile heart and circulation initially
with default parameter values perturbed by only fixed-rate
breathing.
- First, an on-line reduction in lung compliance by a factor of
two. Then, an on-line, 500 ml step increase in the functional
reserve volume of the lungs.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_6.
- Open the file parameters_6 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: waveform: 1 6 15 and
breathing: 1.
- Save the file parameters_6.
- Run the following command at the Linux prompt:
rcvsim parameters_6 foo6
- Some time in the midst of the simulation, type ``p'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Re-assign the the following parameter: Clu: 126.25.
- Save the file parameters_6.
- Type ``r'' followed by
 RETURN
RETURN at standard input.
at standard input.
- At a subsequent time during the simulation, type ``p'' followed
by
 RETURN
RETURN at standard input.
at standard input.
- Re-assign the the following parameter: Qfrs: 500.
- Save the file parameters_6.
- Type ``r'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Execution Output:
- A WAVE window will appear and will automatically scroll through
the simulated data with annotations. The automatic scrolling will
stop once Step 6. is executed. After Steps 7.-9. are executed,
automatic scrolling will resume until Step 10. has been executed.
When Steps 11.-13. are executed, the automatic scrolling will resume
until a total of 300 seconds of data have been simulated.
Figure 14 illustrates the WAVE window during the
reduction in lung compliance. (Note that this reduction does not
alter instantaneous lung volume; see Section 5.3.2).
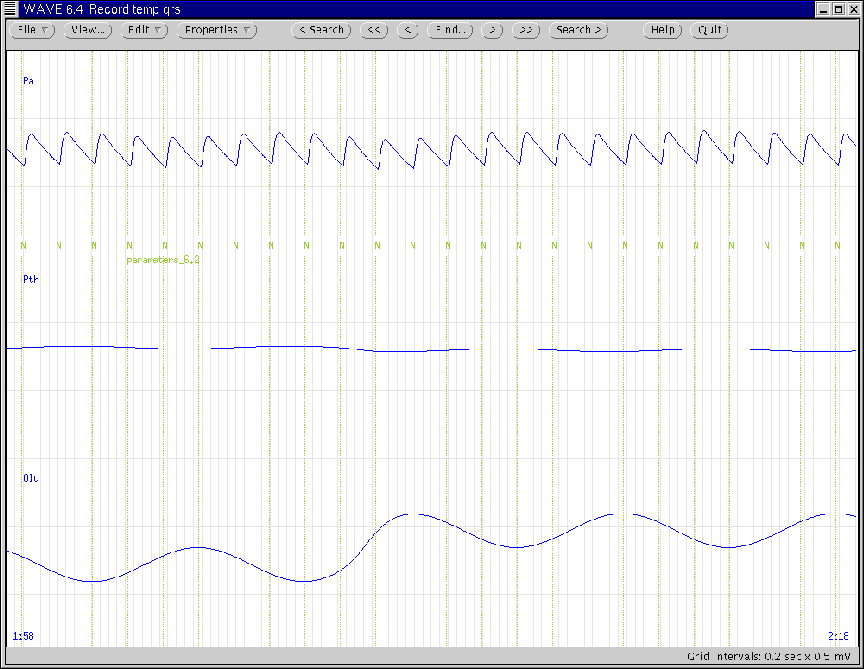
Figure 15 illustrates the WAVE window during the
time of the step increase in the functional reserve volume of the
lungs. (Note that the step increase occurs once the current
respiratory cycle is complete.)
- The following files will be created in the current directory:
foo6.dat, foo6.qrs, foo6.hea, parameters_6.0,
parameters_6.1, and parameters_6.2.
Figure 14:
WAVE window generated according to the first
parameter update of Ex. 6.
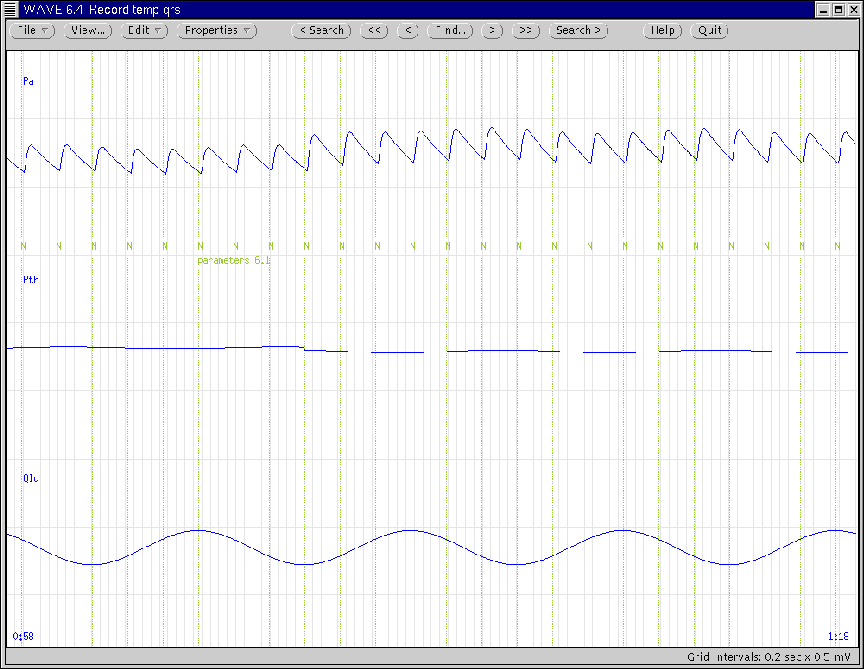 |
Figure 15:
WAVE window generated according to the second
parameter update of Ex. 6.
 |
Ex. 7
- Desired Execution:
- On-line display of systemic arterial pressure and heart rate
with annotations.
- Uncontrolled, unperturbed, intact pulsatile heart and
circulation initially with default parameter values.
- First on-line hemmorhage of 500 ml. Then, on-line initiation of
arterial and cardiopulmonary baroreflex control.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_7.
- Open the file parameters_7 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: waveform: 1 26, baro: 3, bgain: 0, again: 0, and pgain: 0.
- Save the file parameters_7.
- Run the following command at the Linux prompt:
rcvsim parameters_7 foo7
- Some time in the midst of the simulation, type ``p'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Re-assign the the following parameter: Qtot: 4500.
- Save the file parameters_7.
- Type ``r'' followed by
 RETURN
RETURN at standard input.
at standard input.
- At a subsequent time during the simulation, type ``p'' followed
by
 RETURN
RETURN at standard input.
at standard input.
- Re-assign the the following parameters: bgain: 1, again: 1, and pgain: 1.
- Save the file parameters_7.
- Type ``r'' followed by
 RETURN
RETURN at standard input.
at standard input.
- Execution Output:
- A WAVE window will appear and will automatically scroll through
the simulated data with annotations. The automatic scrolling will
stop once Step 6. is executed. After Steps 7.-9. are executed,
automatic scrolling will resume until Step 10. has been executed.
When Steps 11.-13. are executed, the automatic scrolling will resume
until a total of 300 seconds of data have been simulated.
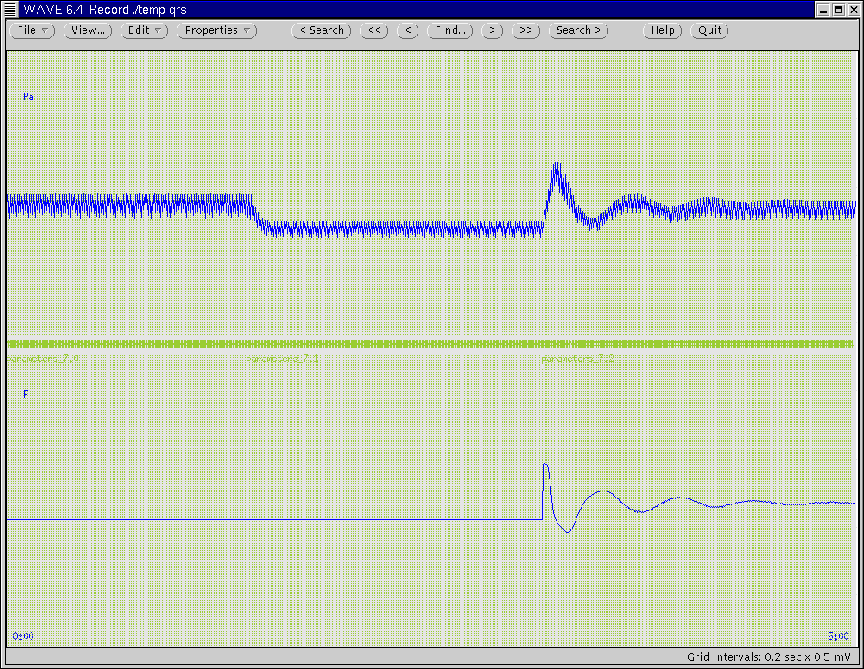
Figure 16 illustrates the WAVE window once the
simulation is complete. The Time variable of the WAVE window
is set such that the waveforms over the entire simulation period may
be viewed all at once. (Note how systemic arterial pressure has
been returned to near normal values with the baroreflex systems.)
- The following files will be created in the current directory:
foo7.dat, foo7.qrs, foo7.hea, parameters_7.0,
parameters_7.1, and parameters_7.2.
Figure 16:
WAVE window generated according to Ex. 7 in which the
time duration of the window has been expanded to illustrate the
entire simulation period.
 |
Ex. 8
- Desired Execution:
- On-line display of a cardiac output curve.
- On-line display of systemic arterial and venous pressure and
left ventricle flow rate.
- Uncontrolled, unperturbed, heart-lung unit preparation with
default parameter values.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_8.
- Open the file parameters_8 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: time: 1000, waveform: 1 2 17, preparation: 1, Pas: 1000, and Pvs: 2.
- Save the file parameters_8.
- Run the following command at the Linux prompt:
rcvsim parameters_8 foo8
- Execution Output:
- A WAVE window will appear and will automatically scroll through
the simulated data as they are being calculated until the entire
cardiac output curve has been measured. Figure 17
illustrates the WAVE window at the end of the simulation. (As is
always the case with viewing waveforms on-line, the parameter values
may be updated prior to simulation termination, if desired.)
- Then, the Gnuplot window of Figure 18 will appear
immediately following the calculation of the entire cardiac output
curve.
- The following files will be created in the current directory:
foo8.dat, foo8.qrs, foo8.hea, foo8.txt and
parameters_8.0.
Figure 17:
WAVE window generated according to Ex. 8.
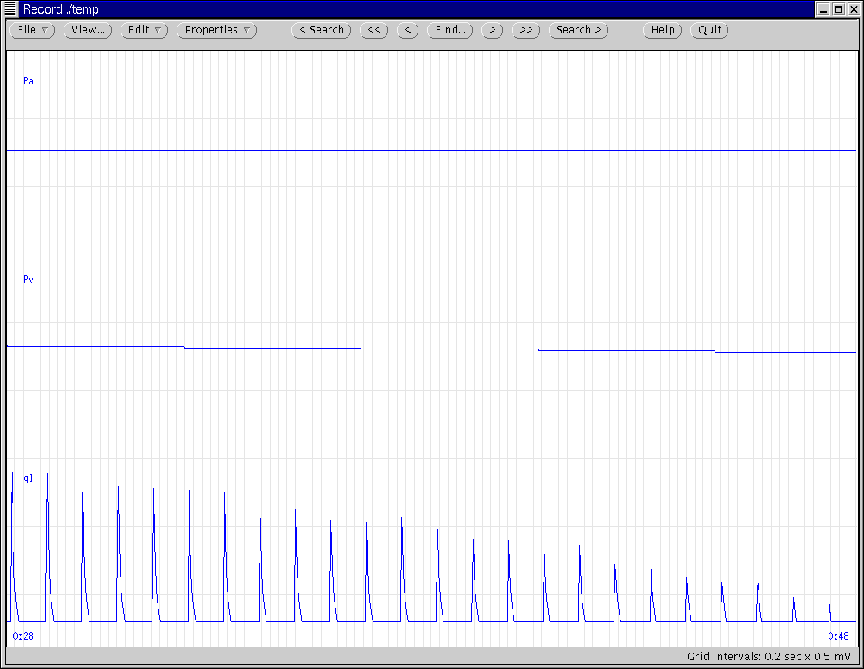 |
Figure 18:
Gnuplot window generated according to Ex. 8, Ex. 10, and
Ex. 11.
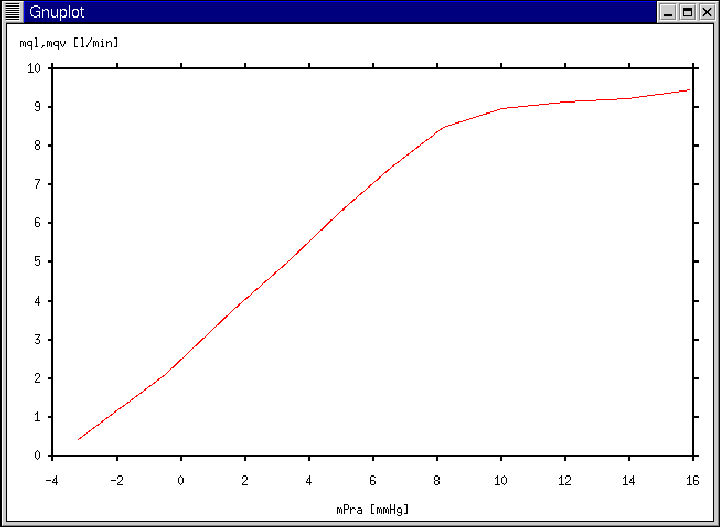 |
Ex. 9
- Desired Execution:
- On-line display of a venous return curve.
- Uncontrolled, unperturbed, systemic circulation preparation with
default parameter values.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_9.
- Open the file parameters_9 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: time: 1000, waveform: -1, preparation: 2, and Crds: 5.
- Save the file parameters_9.
- Run the following command at the Linux prompt:
rcvsim parameters_9 foo9
- Execution Output:
- The Gnuplot window of Figure 19 will appear
immediately following the calculation of the entire venous return
curve.
- The following files will be created in the current directory:
foo9.dat, foo9.qrs, foo9.hea, foo9.txt and
parameters_9.0.
Figure 19:
Gnuplot window generated according to Ex. 9.
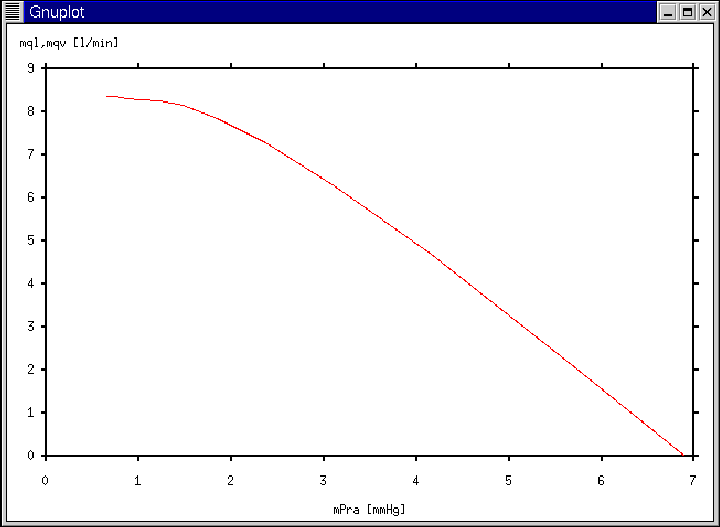 |
Ex. 10
- Desired Execution:
- On-line display of multiple cardiac output and venous return
curves.
- Uncontrolled, unperturbed, heart-lung unit and systemic
circulation preparations with default parameter values and after a
25% increase in systemic arterial resistance, mean systemic
pressure, heart rate, and contractility.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_10.
- Open the file parameters_10 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: time: 1000, waveform: -1, preparation: 1, Pas: 1000, and Pvs: 2.
- Save the file parameters_10.
- Run the following command at the Linux prompt:
rcvsim parameters_10 foo10a
- Once the simulation is complete, re-assign the following
parameters: numerics: 1, preparation: 2, and Crds: 5.
- Save the file parameters_10.
- Run the following command at the Linux prompt:
rcvsim parameters_10 foo10b
- Once the simulation is complete, re-assign the following
parameters: Ra: 1.25 and Pms: 8.625.
- Save the file parameters_10.
- Run the following command at the Linux prompt:
rcvsim parameters_10 foo10c
- Once the simulation is complete, re-assign the following
parameters: preparation: 1, Cls: 0.3, Crs: 0.9,
and F: 1.5.
- Save the file parameters_10.
- Run the following command at the Linux prompt:
rcvsim parameters_10 foo10d
- Execution Output:
- A total of four Gnuplot windows will be displayed beginning with
a window displaying a single cardiac output curve (see
Figure 18); then a window displaying the previous cardiac
output curve with a venous return curve; followed by a window
displaying these two previous curves with another venous return
curve which is enhanced; and ending with a window displaying each of
these three curves and an additional enhanced cardiac output curve
(see Figure 20). (Note the increase in average
cardiac output that occurs due to the enhancement of the curves.)
Each of the four windows will appear immediately after the
completion of each of the four rcvsim executions (Steps 5.,
8., 11., 14.).
- The following files will be created in the current directory:
foo10a.dat, foo10a.qrs, foo10a.hea, foo10a.txt,
foo10b.dat, foo10b.qrs, foo10b.hea, foo10b.txt,
foo10c.dat, foo10c.qrs, foo10c.hea, foo10c.txt,
foo10d.dat, foo10d.qrs, foo10d.hea, foo10d.txt
and parameters_10.0.
Figure 20:
Gnuplot window generated according to Ex. 10 and Ex. 11.
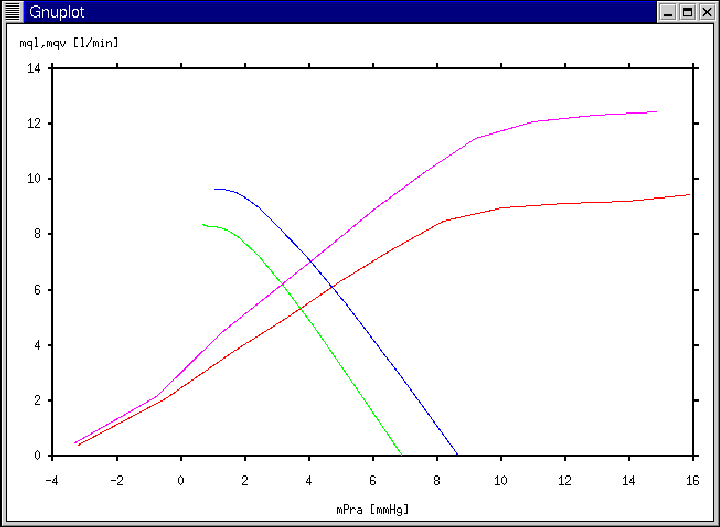 |
Ex. 11
- Desired Execution:
- Off-line display of the cardiac output and venous return curves
generated from Ex. 10.
- Required Steps:
- Change directory to that which contains the simulated data of
Ex. 10.
- Run gnuplot at the Linux prompt.
- At the Gnuplot prompt, enter the following commands:
set tics out
set xlabel 'mPra [mmHg]'
set ylabel 'mql,mqv [l/min]'
plot 'foo10a.txt' using 1:2 notitle with lines
replot 'foo10b.txt' using 1:2 notitle with lines
replot 'foo10c.txt' using 1:2 notitle with lines
replot 'foo10d.txt' using 1:2 notitle with lines
(Note that the plot command is similar to numerics: 0, while
the replot command is analogous to numerics: 1.)
- Execution Output:
- A total of four Gnuplot windows will be displayed beginning with
a window displaying a single cardiac output curve (see
Figure 18); then a window displaying the previous cardiac
output curve with a venous return curve; followed by a window
displaying these two previous curves with another venous return
curve which is enhanced; and ending with a window displaying each of
these three curves and an additional enhanced cardiac output curve
(see Figure 20).
Ex. 12
- Desired Execution:
- On-line display of average cardiac output as a function of
average systemic arterial pressure.
- Uncontrolled, unperturbed, heart-lung unit preparation with
default parameter values.
- Required Steps:
- Copy file $DIR/bin/parameters.def to the current
directory with the new file name parameters_12.
- Open the file parameters_12 with any text editor (e.g.,
emacs).
- Re-assign the following parameters: time: 1000, waveform: -1, numerics: 2, preparation: 1, Pas: 30, and Pvs: 100.
- Save the file parameters_12.
- Run the following command at the Linux prompt:
rcvsim parameters_12 foo12
- Execution Output:
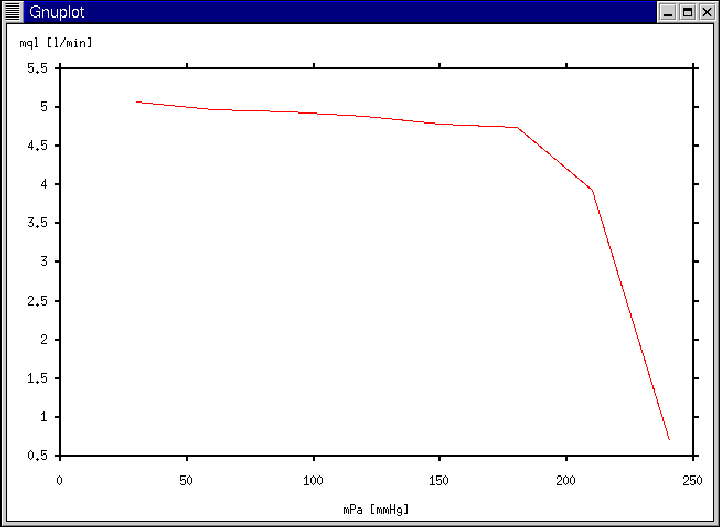
- The Gnuplot window of Figure 21 will appear
immediately following the calculation of the entire curve.
- The following files will be created in the current directory:
foo12.dat, foo12.qrs, foo12.hea, foo12.txt
and parameters_12.0.
Figure 21:
Gnuplot window generated according to Ex. 12 and Ex. 13.
 |
Ex. 13
- Desired Execution:
- Off-line display of the curve of average cardiac output as a
function of average systemic arterial pressure generated from Ex.
12.
- Required Steps:
- Change directory to that which contains the simulated data of
Ex. 12.
- Run gnuplot at the Linux prompt.
- At the Gnuplot prompt, enter the following commands:
set tics out
set xlabel 'mPa [mmHg]'
set ylabel 'mql [l/min]'
plot 'foo12.txt' using 3:2 notitle with lines
- Execution Output:
- The Gnuplot window of Figure 21 will again appear.



Next: A Research Example
Up: Software Execution
Previous: Viewing Cardiac Function and Venous Return Curves
Ramakrishna Mukkamala (rama@egr.msu.edu)
2004-02-03

