[j@localhost Code]$ tach -r ecg -a qrs -F 200 > tac
Let us see how easy is to read the result with Scilab. We call Scilab from the same directory. Now we will read the file base containing the original signals and the file tac that contains the result of our analysis
Now we have two variables in Scilab:
We can plot it:
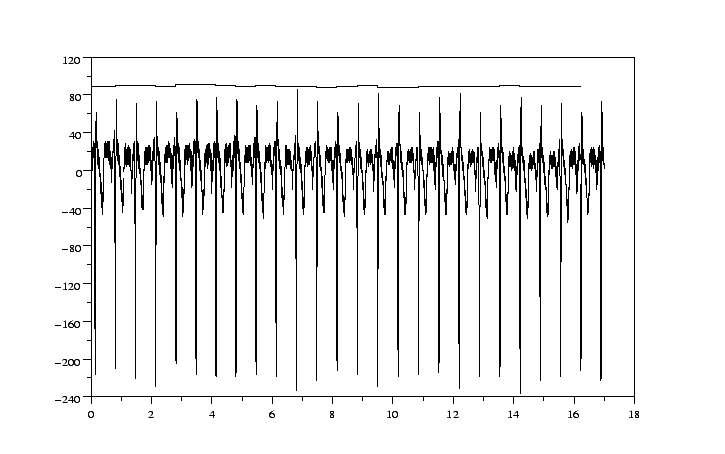
The result can be seen in figure 4.1.
We have corrected the amplitude of the electrocardiographic signal. There is a
good coincidence between the ECG and the heart rate. You can check the result
by eliminating in wave some QRS detections and by observing the changes
with Scilab.
WFDB is very well equipped to analyze electrocardiographic signals. We are
going to make some analysis not implemented in WFDB. We want to detect the
respiratory rate. We have the airway signal in column 14 (13 in WFDB) of
our original recording (base).
To analyze the respiratory rate with Scilab we will use a very simple
approach. Do not pay to much attention to the details since we are using it
only as an example of an alternative analysis made with Scilab.
The approach includes the following steps:
Let us see the code (note that Scilab code is less verbose than the
explanation):
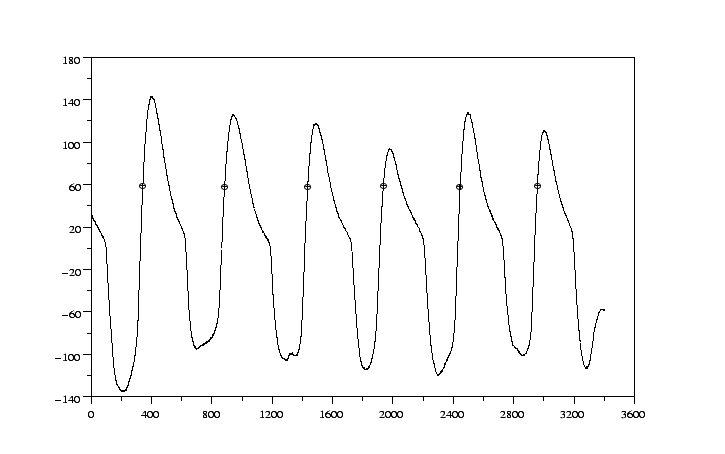
Let us check the result by plotting the detection with the respiratory signal
The result can be seen in figure 4.2.
As we can see, the marked points act as a trigger. But if we want to see the
result in wave, it is not an easy task: we have to create a WFDB
annotation file.
To create an annotation file, our first thought is using wrann. Given
our success by using wrsamp we could try this approach.
Let's see the information about wrann:
So wrann has been conceived to allow editing with a text editor. It
seems a dead end. We would like to have a tool to create annotations easily
from Scilab. We could follow the following approaches:
Having a lot of possibilities increases the probability of error (Do you
remember the simple camera of the introduction?). WFDB is open source
software. Let us explore the code of wrann. After some searching, we
reach to the core of the interface with the external files (lines 111 to 113):
So, it seems that a line is read and unless the first character of the line is
an square bracket it skips the first 9 characters and uses the C function
sscanf to read a long integer (the position of the annotation), a string
(the type) and three integers (the subtype, the chan and the num
field). Probably, knowing the sample, the remainder of the annotation can be
reconstructed. We could try to write some code to emulate the input format of
wrann from Scilab.To do this, we create a string with the format. Let us
see the code (the first lines have been repeated for convenience):
Notice that **dummy** has exactly 9 characters. We choose [15]
because it is an unassigned annotation type. Our creature begins to
live. Now we have to check the result (we return to the terminal):
Isn't it incredible? We introduce garbage and we receive annotations properly
formatted. Let's check that our method functions:
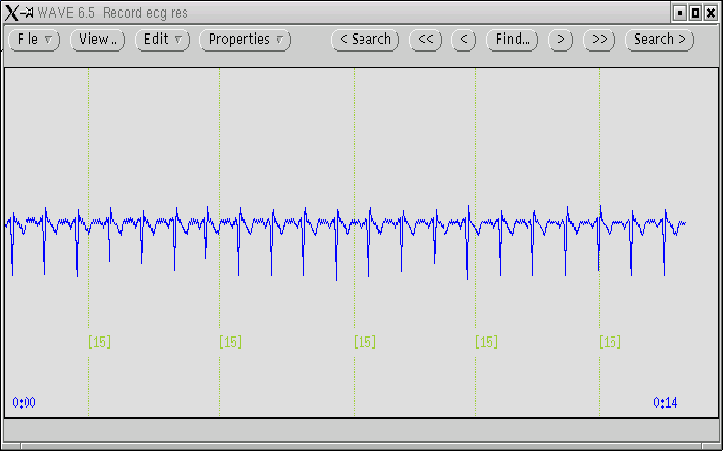
We select the record ecg.dat with the annotation file ecg.res.
The result can be seen in figure 4.3.
After some arrangements, we were able to introduce the result of the
analysis. We have the ECG with the position of the respiratory cycle marked in
the same screen. If we had plotted the respiratory signal (a trivial task), we
could begin a new cycle of editing and exporting results. Our analysis was
trivial (a trigger to detect the respiration cycle), but by using the same
approach you can mark anything in the file of your commercial equipment (if it
exports ASCII files) and edit the result in wave. Let us mention some
things that you can mark:
And many more. Would you not like to see whether the temporal pattern of
spindles is modified by drugs? What is the temporal relation between
K-complexes and spindles? Do really spindles decrease in slow wave sleep or are
they masked by the EEG? Does the auditory evoked potential relate with the
sleep stages? These are questions that you can explore with WFDB software.
Wave uses a very ingenious method for further processing. Let us adapt it to
use Scilab. We define the editor (any other editor can be used) and call
wave:
Then you click on File > Analyze and a Notice appears indicating
that the file wavemenu will be copied in your directory. Once you choose
Copy, you can begin to edit the menu. At the end you add the
following lines:
The main features of the connection are illustrated here:
Now you save the file and click Reread menu in the Analyze
window. A new button called Hello Scilab appears. If you click it, a
small window with a message indicating the file that you are processing
appears.
Do not pay too much atention to this subsection. A lot of things can fail with
this approach: Scilab must be installed, you have to be in the proper
directory, you have to be able to write the file (com.sce) and above all
the synthax is very difficult. It is much better to call scilab from the
command line. This point was included only to show the versatility of
Wave.
In this section we were able to mix the analysis made by external programs with
some elementary analysis made by Scilab. Scilab is a general mathematical
package, Wave is a friendly viewer with a lot of powerful possibilities, WFDB
applications are a set of very well conceived and checked programs for signal
and annotation processing and analysis, including many designed for
electrocardiographic analysis. In this chapter we made them work together by
acting on ASCII files.
[j@localhost Code]$ scilab
===========
S c i l a b
===========
scilab-2.6
Copyright (C) 1989-2001 INRIA
Startup execution:
loading initial environment
-->tac = fscanfMat("tac");
-->size(tac)
ans =
! 3241. 1. !
-->base = fscanfMat("base");
-->size(base)
ans =
! 3400. 19. !
-->time1 = (1:3241)/200;
-->signal1 = tac;
-->time2 = (1:3400)/200;
-->signal2 = base(:,10);
-->plot2d(time1,signal1)
-->plot2d(time2,signal2/5)
Creating some analysis tools with Scilab
-->base = fscanfMat("base"); // reading the file
-->res = base(:,14); // extracting column 14
-->over60 = 1*(res>60); // values over 60
-->changes = over60(2:$)-over60(1:$-1); // changes can be -1, 0 ,1
-->ndx = find( changes >0 ) // detection of values 1
ndx =
! 342. 884. 1435. 1936. 2445. 2957.
-->plot(res) // the respiratory signal
-->plot2d(ndx,res(ndx),-3) // plotting ndx as points
Trying to create an annotation file with the result
The usual application for wrann is as an aid to annotation file
editing: an annotation file may be translated into ASCII format
using rdann, edited using a text editor, and then translated back
into annotation file format using wrann.
while (fgets(line, sizeof(line), stdin) != NULL) {
p = line+9;
if (line[0] == '[')
while (*p != ']')
p++;
while (*p != ' ')
p++;
(void)sscanf(p+1, "%ld%s%d%d%d", &tm, annstr, &sub, &ch, &nm);
annot.anntyp = strann(annstr);
annot.time = tm; annot.subtyp = sub; annot.chan = ch; annot.num = nm;
...
-->base = fscanfMat("base");
-->res = base(:,14);
-->over60 = 1*(res>60);
-->changes = over60(2:$)-over60(1:$-1);
-->ndx = find( changes >0 )
ndx =
! 342. 884. 1435. 1936. 2445. 2957. !
-->format_ann = "**dummy** %13.0f [15] 0 0 0"; // our discovery
-->fprintfMat("dummy_file",ndx',format_ann); // store to a file
[j@localhost Code]$ cat dummy_file
**dummy** 342 [15] 0 0 0
**dummy** 884 [15] 0 0 0
**dummy** 1435 [15] 0 0 0
**dummy** 1936 [15] 0 0 0
**dummy** 2445 [15] 0 0 0
**dummy** 2957 [15] 0 0 0
[j@localhost Code]$ cat dummy_file | wrann -r ecg -a res
[j@localhost Code]$ rdann -r ecg -a res
0:01.710 342 [15] 0 0 0
0:04.420 884 [15] 0 0 0
0:07.175 1435 [15] 0 0 0
0:09.680 1936 [15] 0 0 0
0:12.225 2445 [15] 0 0 0
0:14.785 2957 [15] 0 0 0
[j@localhost Code]$ wave -r ecg -a res
Using Scilab as a tool from wave
[j@localhost Code]$ export EDITOR=/usr/openwin/bin/textedit
[j@localhost Code]$ wave -r 100s
...
# list contains three signals). Adjust the -rx and -rz options to obtain the
# desired viewpoint.
# Add additional entries below.
Hello Scilab echo "key= x_message (\"You are reading record $RECORD\" ) " \
> com.sce; \
echo "exit()" >> com.sce; \
scilab -nw -f com.sce
In summary
![]()
![]()
![]()
![]()
Next: Final considerations
Up: Applying PhysioNet tools to
Previous: Editing the result
Contents
j
2002-12-11