This database is a subset of the data described in
Please cite this publication when referencing this material, and also include the standard citation for PhysioNet:
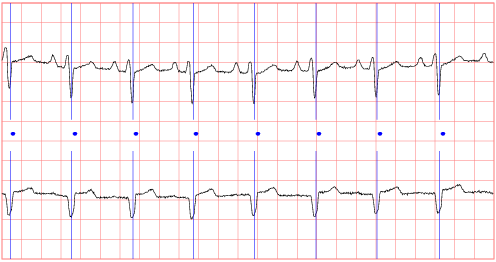
This database includes long-term ECG recordings from 15 subjects (11 men, aged 22 to 71, and 4 women, aged 54 to 63) with severe congestive heart failure (NYHA class 3–4). This group of subjects was part of a larger study group receiving conventional medical therapy prior to receiving the oral inotropic agent, milrinone. Further details about the larger study group are available in the first reference cited above. A number of additional studies have made use of these recordings; see the additional references below.
The individual recordings are each about 20 hours in duration, and contain two ECG signals each sampled at 250 samples per second with 12-bit resolution over a range of ±10 millivolts. The original analog recordings were made at Boston's Beth Israel Hospital (now the Beth Israel Deaconess Medical Center) using ambulatory ECG recorders with a typical recording bandwidth of approximately 0.1 Hz to 40 Hz. Annotation files (with the suffix .ecg) were prepared using an automated detector and have not been corrected manually.
Additional References
- Moody GB, Mark RG, Bump MA, Weinstein JS, Berman AD, Mietus JE, Goldberger AL. Clinical validation of the ECG-derived respiration (EDR) technique. Computers in Cardiology 1986; 13:507–510.
- Goldberger AL, Rigney DR, Mietus J, Antman EM, Greenwald S. Nonlinear dynamics in sudden cardiac death syndrome: heartrate oscillations and bifurcations. Experientia 1988 Dec 1; 44(11–12):983–987. [View Abstract]
- Peng C-K, Mietus J, Hausdorff JM, Havlin S, Stanley HE, Goldberger AL. Long-range anticorrelations and non-Gaussian behavior of the heartbeat. Physical Review Letters 1993 Mar 1; 70(9):1343–1346.
- Peng C-K, Havlin S, Stanley HE, Goldberger AL. Quantification of scaling exponents and crossover phenomena in nonstationary heartbeat time series. Chaos 1995; 5(1):82–87.
- Poon C-S, Merrill CK. Decrease of cardiac chaos in congestive heart failure. Nature 1997 Oct 2; 389(6650):492–495. [View Abstract]
- Ivanov PCh, Bunde A, Amaral LAN, Havlin S, Fritsch-Yelle J, Baevsky RM, Stanley HE, Goldberger AL. Sleep-wake differences in scaling behavior of the human heartbeat: analysis of terrestrial and long-term space flight data. Europhysics Letters 1999; 48(5):594–600.
- Ivanov PCh, Amaral LAN, Goldberger AL, Havlin S, Rosenblum MG, Struzik ZR, Stanley HE. Multifractality in human heart rate dynamics. Nature 1999 Jun 3; 399(6735):461–465. [View Abstract]
- Teich MC, Lowen SB, Jost BM, Vibe-Rhymer K, Heneghan C. Heart rate variability: measures and models. In Nonlinear Biomedical Signal Processing, Vol. II, Dynamic Analysis and Modeling (Ed. M Akay), ch. 6, pp 159–213. New York: IEEE Press, 2001. [The 15 recordings identified in Appendix A of this reference by names of the form annnn are those belonging to this database.]
- Mietus JE, Peng C-K, Henry I, Goldsmith RL, Goldberger AL. The pNNx files: re-examining a widely used heart rate variability measure. Heart 2002; 88:378–380. [View Abstract]
Name Last modified Size Description
Parent Directory -
local.css 28-Oct-2015 22:53 3.1K
DOI 21-Sep-2015 19:00 19
chf01.hea 15-Feb-2012 00:22 148 header file
chf02.hea 15-Feb-2012 00:22 147 header file
chf03.hea 15-Feb-2012 00:22 147 header file
chf04.hea 15-Feb-2012 00:22 148 header file
chf05.hea 15-Feb-2012 00:21 148 header file
chf06.hea 15-Feb-2012 00:21 148 header file
chf07.hea 15-Feb-2012 00:21 147 header file
chf08.hea 15-Feb-2012 00:21 147 header file
chf09.hea 15-Feb-2012 00:21 143 header file
chf10.hea 15-Feb-2012 00:21 146 header file
chf11.hea 15-Feb-2012 00:21 148 header file
chf12.hea 15-Feb-2012 00:21 145 header file
chf13.hea 15-Feb-2012 00:21 149 header file
chf14.hea 15-Feb-2012 00:21 147 header file
chf15.hea 15-Feb-2012 00:21 152 header file
SHA256SUMS 18-Sep-2007 18:37 3.9K
SHA1SUMS 13-Jul-2005 03:57 2.6K
MD5SUMS 13-Jul-2005 03:50 2.2K
chf02.ecg 15-Jul-2003 05:48 224K unaudited annotations
chf04.ecg 15-Jul-2003 04:28 540K unaudited annotations
chf01.hea- 19-Feb-2003 02:33 138 header file
chf02.hea- 19-Feb-2003 02:33 137 header file
chf03.hea- 19-Feb-2003 02:33 137 header file
chf04.hea- 19-Feb-2003 02:33 138 header file
chf05.hea- 19-Feb-2003 02:33 138 header file
chf06.hea- 19-Feb-2003 02:32 138 header file
chf07.hea- 19-Feb-2003 02:32 137 header file
chf08.hea- 19-Feb-2003 02:32 137 header file
chf09.hea- 19-Feb-2003 02:32 133 header file
chf10.hea- 19-Feb-2003 02:32 136 header file
chf11.hea- 19-Feb-2003 02:32 138 header file
chf12.hea- 19-Feb-2003 02:32 135 header file
chf13.hea- 19-Feb-2003 02:32 139 header file
chf14.hea- 19-Feb-2003 02:31 137 header file
chf15.hea- 19-Feb-2003 02:31 142 header file
ANNOTATORS 13-Oct-2000 20:29 58 list of annotators
RECORDS 06-Oct-2000 05:58 90 list of record names
chf15.ecg 06-Oct-2000 05:57 225K unaudited annotations
chf14.ecg 06-Oct-2000 05:56 183K unaudited annotations
chf13.ecg 06-Oct-2000 05:55 226K unaudited annotations
chf12.ecg 06-Oct-2000 05:55 225K unaudited annotations
chf11.ecg 06-Oct-2000 05:54 226K unaudited annotations
chf10.ecg 06-Oct-2000 05:52 288K unaudited annotations
chf09.ecg 06-Oct-2000 05:52 225K unaudited annotations
chf08.ecg 06-Oct-2000 05:50 177K unaudited annotations
chf07.ecg 06-Oct-2000 05:50 181K unaudited annotations
chf06.ecg 06-Oct-2000 05:49 233K unaudited annotations
chf05.ecg 06-Oct-2000 05:47 233K unaudited annotations
chf03.ecg 06-Oct-2000 05:43 159K unaudited annotations
chf02.ecg- 06-Oct-2000 05:42 222K unaudited annotations
chf01.ecg 06-Oct-2000 05:39 148K unaudited annotations
chf15.dat 06-Oct-2000 05:17 51M digitized signal(s)
chf14.dat 06-Oct-2000 05:14 51M digitized signal(s)
chf13.dat 06-Oct-2000 05:13 51M digitized signal(s)
chf12.dat 06-Oct-2000 05:11 51M digitized signal(s)
chf11.dat 06-Oct-2000 05:10 51M digitized signal(s)
chf10.dat 06-Oct-2000 05:09 51M digitized signal(s)
chf09.dat 06-Oct-2000 05:04 51M digitized signal(s)
chf08.dat 06-Oct-2000 05:03 51M digitized signal(s)
chf07.dat 06-Oct-2000 05:01 51M digitized signal(s)
chf06.dat 06-Oct-2000 05:00 51M digitized signal(s)
chf05.dat 06-Oct-2000 04:58 51M digitized signal(s)
chf04.dat 06-Oct-2000 04:57 51M digitized signal(s)
chf04.ecg- 06-Oct-2000 04:56 173K unaudited annotations
chf03.dat 06-Oct-2000 04:55 51M digitized signal(s)
chf02.dat 06-Oct-2000 04:54 51M digitized signal(s)
chf01.dat 06-Oct-2000 04:53 51M digitized signal(s)
|
If you would like help understanding, using, or downloading content, please see our Frequently Asked Questions. If you have any comments, feedback, or particular questions regarding this page, please send them to the webmaster. Comments and issues can also be raised on PhysioNet's GitHub page. Updated Friday, 28-Oct-2016 22:58:42 CEST |
PhysioNet is supported by the National Institute of General Medical Sciences (NIGMS) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) under NIH grant number 2R01GM104987-09.
|